SPPAS implements an Application Programming Interface (API), with name anndata, to deal with annotated files.
anndata is a free and open source Python library to access and search data from annotated data of the supported formats (xra, TextGrid, eaf…). It can either be used with the Programming Language Python 2.7 or Python 3.7+.
This API allows converting file formats like Elan's EAF, Praat’s TextGrid
and others into a sppasTranscription object and convert
this object into any of these formats. This object allows unified access
to linguistic data from a wide range sources.
This page includes exercises. The script's solutions are
included in the package folder documentation/scripting_solutions.
anndata, an API to manage annotated data
Overview
We are now going to write Python scripts using the anndata API included in SPPAS. This API is useful to read/write and manipulate files annotated from various annotation tools like SPPAS, Praat or Elan.
First of all, it is important to understand the data structure included in the API to be able to use it efficiently.
Why developing a new API?
In the Linguistics field, multimodal annotations contain information
ranging from general linguistic to domain-specific information. Some are
annotated with automatic tools, and some are manually annotated. In
annotation tools, annotated data are mainly represented in the form of
tiers
or tracks
of annotations. Tiers are mostly series of
intervals defined by:
- two time points to represent the beginning and the end of the interval;
- a label to represent the annotation itself.
Of course, depending on the annotation tool, the internal data
representation and the file formats are different. In Praat, tiers can
be represented either by a single point in time
(such tiers are named PointTiers) or two (IntervalTiers) In Elan, points
are not supported; but contrariwise to Praat, unlabelled intervals are
not represented nor saved.
The anndata API was designed to be able to manipulate all data in the same way, regardless of the file type. It supports merging data and annotations from a wide range of heterogeneous data sources.
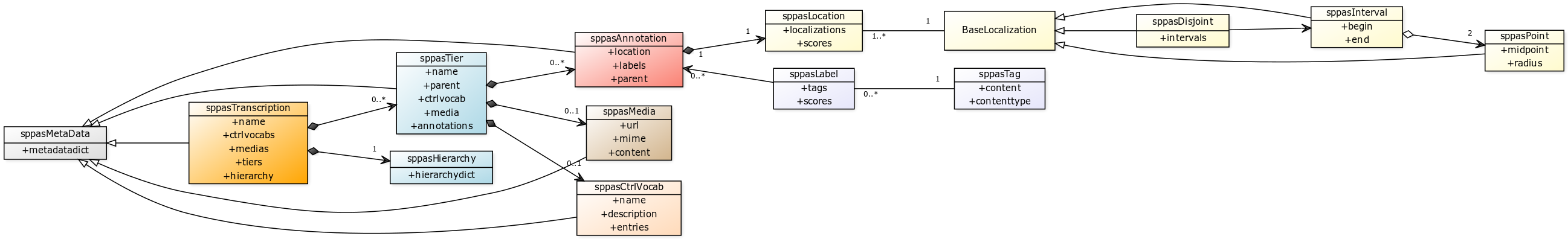
The anndata
API class
diagram
After opening/loading a file, its content is stored in a
sppasTranscription object. A
sppasTranscription has a name, and a list of
sppasTier objects. Tiers can’t share the same name, the
list of tiers can be empty, and a hierarchy between tiers can be
defined. Actually, subdivision relations can be established between
tiers. For example, a tier with phonemes is a subdivision reference for
syllables, or for tokens; and tokens are a subdivision reference for the
orthographic transcription in IPUs. Such subdivisions can be of two
categories: alignment or association.
A sppasTier object has a name, and a list of
sppasAnnotation objects. It can also be associated to a
controlled vocabulary, or a media.
Al these objects contain a set of meta-data.
An annotation is made of two objects:
- a
sppasLocationobject, - a list of
sppasLabelobjects.
A sppasLabel object is representing the content
of the annotation. It is a list of sppasTag each one
associated to a score.
A sppasLocation is representing where this annotation
occurs in the media. Then, a sppasLocation is made of a
list of localization each one associated with a score. A localization is
one of:
- a
sppasPointobject; or - a
sppasIntervalobject, which is made of 2sppasPointobjects; or - an un-used
sppasDisjointobject which is a list ofsppasInterval.
Label representation
Each annotation holds a series of 0 to N labels. A label is also an object made of a list of sppasTag, each one with a score. A sppasTag is mainly represented in the form of a string, freely written by the annotator, but it can also be a boolean (True/False), an integer, a floating number, a point with (x, y) coordinates with an optional radius or a rectangle with (x, y, w, h) coordinates with an optional radius value.
Location representation
In the anndata API, a sppasPoint is considered
as an imprecise value. It is possible to characterize a point
in a space immediately allowing its vagueness by using:
- a midpoint value (center) of the point;
- a radius value.
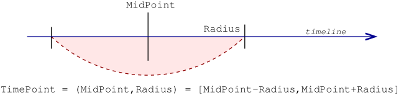
Example
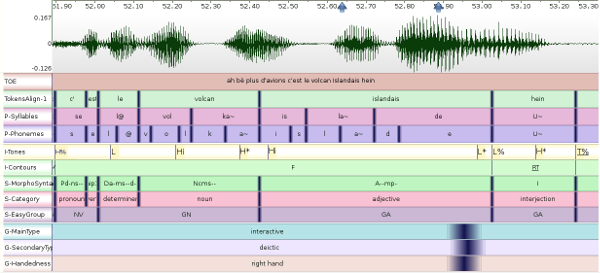
The screenshot below shows an example of multimodal annotated data,
imported from three different annotation tools. Each sppasPoint
is represented by a vertical dark-blue line with a gradient color to
refer to the radius value.
In the screenshot the following radius values were assigned:
- 0ms for prosody,
- 5ms for phonetics, discourse and syntax
- 40ms for gestures.
Creating scripts with anndata
Preparing the data
To practice, you have first to create a new folder in your computer,
on your 'Desktop' for example; with name sppasscripts for example,
and to execute the python IDLE.
Open a File Explorer window and go to the SPPAS folder location.
Then, copy the sppas directory into the newly created
sppasscripts
folder. Then, go to the solution directory and
copy/paste the files skeleton-sppas.py and
F_F_B003-P9-merge.TextGrid into your sppasscripts
folder. Then, open the skeleton script with the python IDLE and execute
it. It will do... nothing! But now, you are ready to do something with the
API of SPPAS!
When using the API, if something forbidden is attempted, the object will raise an Exception which means the program will stop.
Read/Write annotated files
We are being to Open/Read an annotated file of any format (XRA,
TextGrid, Elan, …) and store it into a sppasTranscription
object instance. Then, the object will be saved into another file.
# Create a parser object then parse the input
file.
parser =
sppasRW(input_filename)
trs = parser.read()
# Save the sppasTranscription object into
a file.
parser.set_filename(output_filename)
parser.write(trs)Only these few lines of code are required to convert a file from a format to another one! The appropriate parsing system is extracted from the extension of file name.
To get the list of accepted extensions that the API can read, just
use parser.extensions_in(). The list of accepted extensions
that the API can write is given by
parser.extensions_out().
Practice: Write a script to convert a TextGrid file into CSV (solution: ex10_read_write.py)
Manipulating a sppasTranscription object
The most useful functions to manage the tiers of a sppasTranscription object are:
create_tier()to create an empty tier and to append it,append(tier)to add a tier into the sppasTranscription,pop(index)to remove a tier of the sppasTranscription,find(name, case_sensitive=True)to find a tier from its name.
Below is a piece of code to browse through the list of tiers:
for tier
in trs:
# below, do something with the
tier:
print(tier.get_name())
# Search for a specific tier,
# None is returned if not
found.
phons_tier = trs.find("PhonAlign")Practice: Write a script to select a set of tiers of a file and save them into a new file (solution: ex11_transcription.py).
Manipulating a sppasTier object
A tier is made of a name, a list of annotations, and optionally a
controlled vocabulary and a media. To get the name of a tier, or to fix
a new name, the easier way is to use tier.get_name(). The
following block of code allows getting a tier and changing its name.
# Get the first
tier, with index=0
tier = trs[0]
print(tier.get_name())
tier.set_name("NewName")
print(tier.get_name())The most useful functions to manage annotations of a
sppasTier object are:
create_annotation(location, labels)to create and add a new annotationappend(annotation)to add a new annotation at the end of the listadd(annotation)to add a new annotationpop(index)to delete the annotation of a given indexremove(begin, end)to remove annotations of a given localization rangeis_disjoint(),is_interval(),is_point()to know the type of locationis_string(),is_int(),is_float(),is_bool(),is_fuzzypoint(),is_fuzzyrect()to know the type of labelsfind(begin, end)to get annotations in a given localization rangeget_first_point(),get_last_point()to get respectively the point with the lowest or highest localizationset_radius(radius)to fix the same vagueness value to each localization point
Practice: Write a script to open an annotated file and print information about tiers (solution: ex12_tiers_info.py)
Manipulating a sppasAnnotation object
An annotation is a container for a location and optionally a list of labels. It can be used to manage the labels and tags with the following methods:
is_labelled()returns True if at least asppasTagexists and is not Noneappend_label(label)to add a label at the end of the list of labelsget_labels_best_tag()returns a list with the best tag of each labeladd_tag(tag, score, label_index)to add a tag into a labelremove_tag(tag, label_index)to remove a tag of a label
An annotation object can also be copied with the method
copy(). The location, the labels and the metadata are all
copied; and the id
of the returned annotation is then the same.
It is expected that each annotation of a tier as its own id
, but
the API doesn’t check this.
Practice: Write a script to print information about annotations of a tier (solution: ex13_tiers_info.py)
Search in annotations: Filters
Overview
This section focuses on the problem of searching and retrieving data from annotated corpora.
The filter implementation can only be used together with the
sppasTier() class. The idea is that each
sppasTier() can contain a set of filters that each reduce
the full list of annotations to a subset.
SPPAS filtering system proposes two main axes to filter such data:
- with a boolean function based either on the content, the duration or on the time of annotations,
- with a relation function between annotation locations of two tiers.
A set of filters can be created and combined to get the expected
result. To be able to apply filters to a tier, some data must be loaded
first. First, a new sppasTranscription() has to be created
when loading a file. Then, the tier(s) to apply filters on must be
fixed. Finally, if the input file was NOT an XRA, it is widely
recommended to fix a radius value before using a relation filter.
f =
sppasFilter(tier)When a filter is applied, it returns an instance of
sppasAnnSet which is the set of annotations matching with
the request. It also contains a value
which is the list of
functions that are truly matching for each annotation. Finally,
sppasAnnSet objects can be combined with the operators
|
and &
, and expected to a sppasTier
instance.
Filter on the tag content
The following matching names are proposed to select annotations:
exact
: means that a tag is valid if it strictly corresponds to the expected pattern;contains
means that a tag is valid if it contains the expected pattern;startswith
means that a tag is valid if it starts with the expected pattern;endswith
means that a tag is valid if it ends with the expected pattern.regexp
to define regular expressions.
All these matches can be reversed, to represent does not exactly
match, does not contain, does not start with or does not end with.
Moreover, they can be case-insensitive by adding i
at the
beginning like iexact
, etc. The full list of tag matching
functions is obtained by invoking
sppasTagCompare().get_function_names().
The next examples illustrate how to work with such a pattern-matching
filter. In this example, f1 is a filter used to get all
phonemes with the exact label a
. On the other side,
f2 is a filter that ignores all phonemes matching with
a
(mentioned by the symbol ~
) with a case-insensitive
comparison (iexact means insensitive-exact).
tier = trs.find("PhonAlign")
f = sppasFilter(tier)
ann_set_a = f.tag(exact='a')
ann_set_aA = f.tag(iexact='a')The next example illustrates how to write a complex request. Notice that r1 is equal to r2, but getting r1 is faster:
tier = trs.find("TokensAlign")
f = sppasFilter(tier)
r1 = f.tag(startswith="pa",
not_endswith='a', logic_bool="and")
r2 = f.tag(startswith="pa")
& f.tag(not_endswith='a')With this notation in hands, it is easy to formulate queries, like for
example: Extract words starting by ch
or sh
:
result = f.tag(startswith="ch") |
f.tag(startswith="sh")Practice:: Write a script to extract phonemes /a/ then phonemes /a/, /e/, /A/ and /E/. (solution: ex15_annotation_label_filter.py).
Filter on the duration
The following matching names are proposed to select annotations:
lt
means that the duration of the annotation is lower than the given one;le
means that the duration of the annotation is lower or equal than the given one;gt
means that the duration of the annotation is greater than the given one;ge
means that the duration of the annotation is greater or equal than the given one;eq
means that the duration of the annotation is equal to the given one;ne
means that the duration of the annotation is not equal to the given one.
The full list of duration matching functions is obtained by invoking
sppasDurationCompare().get_function_names().
Next example shows how to get phonemes during between 30 ms and 70 ms. Notice that r1 and r2 are equals!
tier = trs.find("PhonAlign")
f = sppasFilter(tier)
r1 = f.dur(ge=0.03) & f.dur(le=0.07)
r2 = f.dur(ge=0.03, le=0.07, logic_bool="and")Practice: Extract phonemes
aoreduring more than 100ms (solution: ex16_annotation_dur_filter.py).
Filter on position in time
The following matching names are proposed to select annotations:
- rangefrom allows fixing the 'begin' time value,
- rangeto allows fixing the 'end' time value.
Next example allows extracting phonemes a
of the 5 first
seconds:
tier = trs.find("PhonAlign")
f = sppasFilter(tier)
result = f.tag(exact='a') & f.loc(rangefrom=0.,
rangeto=5., logic_bool="and")Creating a relation function
Relations between annotations is crucial if we want to extract multimodal data. The aim here is to select intervals of a tier depending on what is represented in another tier.
James Allen, in 1983, proposed an algebraic framework named Interval Algebra (IA), for qualitative reasoning with time intervals where the binary relationship between a pair of intervals is represented by a subset of 13 atomic relations, that are:
distinct because no pair of definite intervals can be related by more than one of the relationships;
exhaustive because any pair of definite intervals are described by one of the relations;
qualitative (rather than quantitative) because no numeric time spans are considered.
These relations and the operations on them form Allen’s Interval
Algebra
.
Pujari, Kumari and Sattar proposed INDU in 1999: an Interval & Duration network. They extended the IA to model qualitative information about intervals and durations in a single binary constraint network. These duration relations are greater, lower and equal. INDU comprises 25 basic relations between a pair of two intervals.
anndata implements the 13 Allen interval relations:
before, after, meets, met by, overlaps, overlapped by, starts, started
by, finishes, finished by, contains, during and equals; and it also
contains the relations proposed in the INDU model. The full list of
matching functions is obtained by invoking
sppasIntervalCompare().get_function_names().
Moreover, in the implementation of anndata, some
functions accept options:
beforeandafteraccept amax_delayvalue,overlapsandoverlappedbyaccept anoverlap_minvalue and a booleanpercentwhich defines whether the value is absolute or is a percentage.
The next example returns monosyllabic tokens and tokens that are overlapping a syllable (only if the overlap is during more than 40 ms):
tier = trs.find("TokensAlign")
other_tier = trs.find("Syllables")
f = sppasFilter(tier)
f.rel(other_tier, "equals", "overlaps", "overlappedby", min_overlap=0.04)Below is another example of implementing a request. Which syllables stretch across two words?
# Get tiers from a sppasTranscription
object
tier_syll = trs.find("Syllables")
tier_toks = trs.find("TokensAlign")
f = sppasFilter(tier_syll)
# Apply the filter with the relation
function
ann_set = f.rel(tier_toks, "overlaps", "overlappedby")
# To convert filtered data into a
tier:
tier = ann_set.to_tier("SyllStretch")Practice 1: Create a script to get tokens followed by a silence. (Solution: ex17_annotations_relation_filter1.py).
Practice 2: Create a script to get tokens preceded by OR followed by a silence. (Solution: ex17_annotations_relation_filter2.py).
Practice 3: Create a script to get tokens preceded by AND followed by a silence. (Solution: ex17_annotations_relation_filter3.py).
More with SPPAS…
In addition to anndata, SPPAS contains several other API. They are all free and open source Python libraries, with a documentation and a set of tests.
Among others:
- audiodata to manage digital audio data: load, get information, extract channels, re-sample, search for silences, mix channels, etc.
- calculus to perform some math on data, including descriptive statistics.
- resources to access and manage linguistic resources like lexicons, dictionaries, etc.
